|
|
|
|
 |
| Communicating
in Words |
 |
|
|
|


|
|
| GENES THAT ARE ESSENTIAL
FOR SPEECH |
|
First
discovered in the KE
family, several of whose members had specific language
impairments, the FOXP2 (short for “forkhead box P2”)
gene is the first gene that scientists ever associated with
the human ability to speak.
The exact problems caused by mutations in this gene remain
hard to identify , which is not surprising when you
consider the family of genes to which this one belongs.The
FOX family of genes are transcription factors, which means
that they produce proteins that can regulate the expression
of a number of other genes by binding directly to their
DNA. (The binding ability of these particular proteins
comes from their forked shape, from which the gene family
gets its name.) |
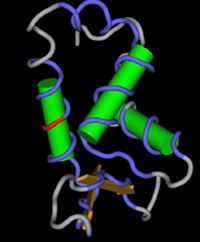
General
form of the FOXP2 protein. The red segment marks the
location of the mutation that caused the specific language
impairments in members of the KE family.
Source: Dr. Simon Fisher |
|
The FOXP2 gene would appear to play an important
role in orchestrating the establishment
of the neural pathways during embryonic development. And in
fact, this gene is extremely well preserved phylogenetically: the
protein that it produces is almost identical in mice and in primates,
which are separated by some 130 million years of evolution.
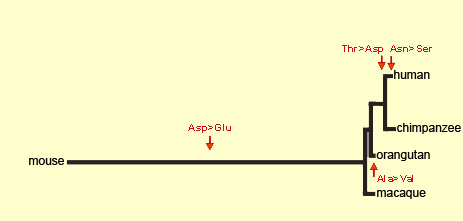
Source: Dr. Svante Pääbo
The protein that the FOXP2 gene produces
in humans differs by only two or three amino acids from the protein
that it produces in other species. It is very likely these two
or three amino acids that make the difference between animals that
cannot speak and humans who can. Moreover, the mutations that caused
this difference are estimated to have occurred between 100 000
and 200 000 years ago, roughly the time that articulate language
first emerged in human beings.
By performing a detailed analysis of the
defective FOXP2 gene sequence in several members of the KE family,
scientists were also able to identify the precise site of the mutation
that caused this gene to malfunction in these individuals. This
mutation occurs on exon 14 of the FOXP2 gene, when the guanine
in a nucleotide is
replaced by an adenine. As it happens, the part of the gene where
this mutation occurs is precisely the one that codes for the “forkhead” portion
of the protein—the part that binds to the DNA on other genes.
This change in a single nucleotide on the FOXP2 gene has a direct
impact on this protein, causing the amino acid arginine to be replaced
with a histidine.
In the hundreds of normal subjects tested, the protein produced
by FOXP2 always has an arginine at this particular site, while
in the members of the KE family who suffered from specific language
impairments, it always had a histidine. Hence there is not a shadow
of a doubt about the mutation that causes this disorder. That said,
it is still amazing to think that the mutation of a single one
of the 2 500 nucleic bases in the FOXP2 gene is sufficient to impair
so vital a faculty as language!
Just around the end
of World War II, the Italian population geneticist Luca Cavalli-Sforza
began constructing genealogical trees that established relationships
among populations throughout the world. This ambitious project
supported certain hypotheses that broke fertile ground for
further research. For example, by cross-tabulating data on
several dozen genes, Cavalli-Sforza established a relationship
between American Indians and Asians. This finding is consistent
with the most common theory about how the New World was populated:
by peoples who crossed from Siberia to Alaska when the Bering
Strait was frozen over during the last great Ice Age, some
30 000 years ago.
Cavalli-Sforza’s findings assumed even greater importance
when he correlated them with analogous studies on languages.
When Cavalli-Sforza compared the genealogical
trees established by geneticists with those established by
linguists, he got some amazing results: with just a few
exceptions, the people who speak each of the 15 major families
of languages are genetically related as well. The obvious explanation
for this remarkable concordance is, of course, that when a
population migrates to a new territory, it takes its genes
along as well as its language.
But there have been many criticisms of Cavalli-Sforza’s
approach, and in particular his way of defining a population.
In the works of Cavalli-Sforza and his followers, the first
step is to define a population, by linguistic criteria among
others. Correlations are then established between these populations
and their languages, which seems like a dangerously circular
approach. It has also been noted that these studies are more
convincing on a small scale or a large scale, but far less
so on an intermediate one. The reason is that it is easier
to distinguish Inuit from Bantu, for example, than to differentiate
the various populations that speak Bantu languages.
In addition, the DNA samples used in many studies come from
blood banks, and the accompanying records may be biased or
false, because for various reasons, when people give blood,
they may report their ethnicity as different from what it actually
is. Once again we see the risks that errors may be found when
controls are applied to both linguistic and genetic data.
|
|
|






